Presentation
PackMem is a computational tool to characterize and quantify interfacial voids in MD simulations of lipid membranes. The analysis of the structural organization of lipid bilayers is generally performed across the normal of the bilayer. In contrast, PackMem performs a topographic analysis of the bilayer surface. Using a Cartesian grid, it allows identifying surface regions defined as lipid-packing defects where lipids are loosely packed, thereby leading to cavities where aliphatic carbons are exposed to the solvent either deep or close to the membrane surface. Thus, PackMem provides a detailed and quantitative characterization of membrane interfacial regions in term of lipid-packing properties.
Deep/Shallow
Deep and Shallow defects are defined as follows:
- Deep: grid cells corresponding to voids where aliphatic atoms are deeper than d Å below the central atom of glycerol and are vertically accessible from the outside.
- Shallow: grid cells corresponding to accessible aliphatic atoms that are less than d Å below the central atom of glycerol and are vertically accessible from the outside.
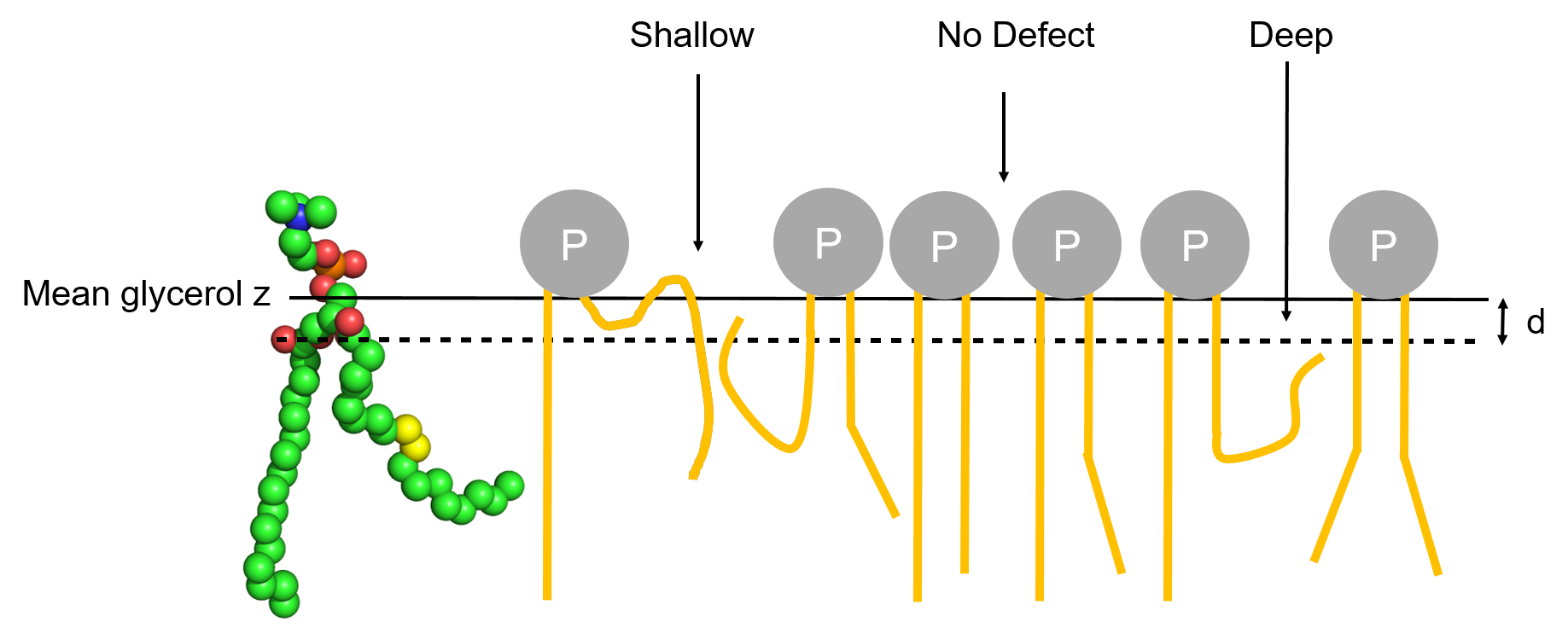
Schematic representation of the two types of lipid-packing defects.
Algorithm
The driving idea of the algorithm is to map the membrane surface according to a grid parallel to the plane of the bilayer. Looking at the grid from above, we assign a value of 0 to each grid cell where the first atom vertically met is an aliphatic atom. Conversely, we assign a value of 1 to all other cells, that is cells where the first atom met vertically is a polar atom. In our analysis, only lipids are considered (water, ions or any other molecules are ignored from the analysis) and each leaflet is treated separately. We opted to loop over all atoms and check which grid cells overlap with each atom. In details, the algorithm starts by constructing a grid of 1 Å x 1 Å resolution and follows these steps in pseudo-code:
initialize a matrix M (representing the grid) with 0
for atom in all lipid atoms of one leaflet
if atom z coordinate is below d Å of the central glycerol atom
of the same lipid
go to next iteration
# the grid cells overlapping the atom
# (M[i,j]) stay at the same value
else
if atom is aliphatic
add 0.001 to the grid cells (M[i,j]) overlapping the atom
else
# the atom is polar
add 1 to the grid cells (M[i,j]) overlapping the atom
endif
endif
endfor
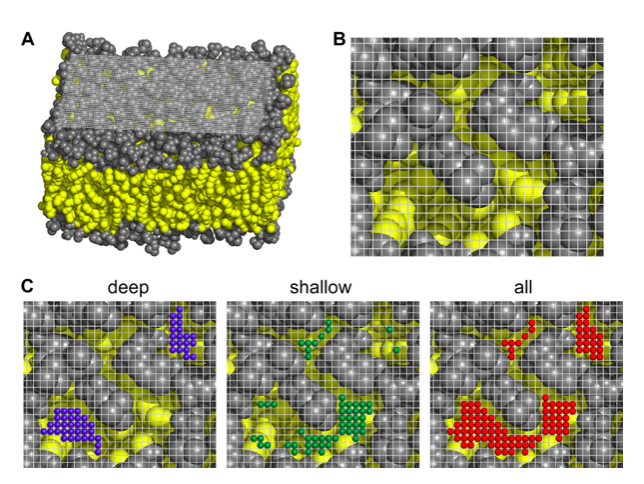
(A,B) Views of the cartesian grid used to spot packing defect on the membrane surface. (C) Examples of the different types of lipid-packing defects found on a membrane. The polar heads are colored in grey and the apolar ones in yellow. The grid is also shown. The small colored balls are the elementary defects found.
